Endothelial cells take pivotal roles in the heart and the vascular system and their differentiation, subspecification and function is determined by cell type- and organ-specific gene expression (Lother & Bergemann et al. 2018).
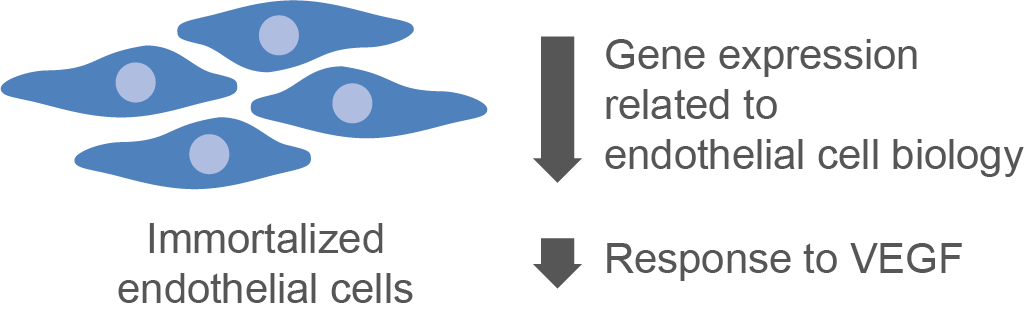
A stable, in vitro cardiac endothelial cell line could provide high cell numbers as needed for many epigenetic analyses and facilitate the understanding of molecular mechanisms involved in endothelial cell biology. To test their suitability for transcriptomic or epigenetic studies, we compared the transcriptome of cultured immortalized mouse cardiac endothelial cells (MCEC) to primary cardiac endothelial cells (pEC). However, in MCEC we found a broad downregulation of genes that are highly expressed in pEC, including well-described markers of endothelial cell differentiation. Accordingly, systematic analysis revealed a downregulation of genes associated with typical endothelial cell functions in MCEC, while genes related to mitotic cell cycle were upregulated when compared to pEC. In conclusion, the findings from this study suggest that primary cardiac endothelial cells should preferably be used for genome-wide transcriptome or epigenome studies. The suitability of in vitro cell lines for experiments investigating single genes or signaling pathways should be carefully validated before use.
Deng et al. Gene expression in immortalized versus primary isolated cardiac endothelial cells. Scientific Reports. 2020;10:1-9
